With the assistance of 3-D glasses and a projection screen, biologists are now able to study their slides as if they were standing directly on the
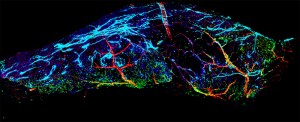
glass itself. Associate professor of electrical and computer engineering Andrew Cohen led the development of the innovative software that makes this possible.
The software was crafted to analyze data biologists have collected using 3-D microscopes. Using the same type of 3-D gaming hardware many use recreationally, Cohen and his team created a cutting edge visual program that allows biologists to see what they’re studying in five dimensions: The x, y and z planes, plus color and time.
“We want to find the knobs and switches that control how the cells behave, ideally, and although it sounds like a biology problem, I think of it very much as an engineering problem,” Cohen said.
The team uses a software program they developed called LEVER — Lineage Editing and Validation program — to track and tag the cells they are interested in. After the equations have been performed on the cell images, the user can identify the exact point the cells are dividing and zoom in on their area of interest, obtaining a spatial understanding not possible looking through a microscope.
“Now you can watch the cells’ movement over and over again, inside the space,” Eric Wait, a doctoral researcher in Cohen’s lab, said.
The software enables scientists to look at cells more comprehensively. Slipping on the 3-D glasses allows one to see how close the cells exist in relation to each other and exactly where they touch and divide. Think of it as the biological kin to the added clarity software maps allow when zooming in on a specific geographic location.
The new visual perspective has been helping biologists in Albany, New York, at the Neural Stem Cell Institute that Cohen has been collaborating with since 2005. Chris Bjornsson, a research scientist at the institute, provides Cohen’s lab with the data the institute collects. He is part of a project that studies neurogenesis — using this 3-D software to study how the stem cells’ environments impact their growth, development and aging.
“Stem cells have a lot of promise for practical application for spinal cord injuries, for neurodegenerative disease, but in order to use them as a treatment you really would like to understand how they work,” Cohen said.
“Really there’s no way to visualize these cells or get any quantitative data out of our studies without using the software these guys have developed,” Bjornsson said.
The program is still in its beginning stages but holds the potential to lead to great therapeutic advancements.
“Many of the same mathematical techniques we use to characterize a computer will also characterize exactly how stem cells work,” Cohen continued.
“The idea would be, once we have our tools sharpened and we can get data accurately, then we can do some problematic analysis,” Wait said. “Now that we know we can get this data, let’s take a bunch of different tests and start discovering why these things happen. Is it actually something the stem cells do? Or is it something that the environment does instead? Once we figure that out then it would go to therapeutics.”
Though the program thus far has been most specifically adapted to study stem cells, the software became free and open source Oct. 10 and many local institutes are currently beginning the process toward using LEVER for their own studies.
“We should be able to extend this tool more generally without much trouble,” Wait said.
Currently, Thomas Jefferson University is also beginning to utilize the program to better study clathrin, a protein that acts as a gateway between a cell and its environment and is often responsible for the admittance of pathogens into the cell.
James Keen, professor of biochemistry and molecular biology at Thomas Jefferson, will work with the programmers to collect data for his research on clathrin.
“The tools that they have developed are very powerful,” Keen said. “We’re trying to follow the clathrin inside the cell. If we can understand that we can understand a lot about not [only] how viruses are processed in cells, but also how viruses respond to growth signals or misrespond to growth signals.”
The time tracking feature would serve other research as well as Keen’s. For instance, biologists at The Children’s Hospital of Philadelphia are interested in using the program to study scratch assays, which are used to research the wound healing process. Also in the Philadelphia community, the University of Pennsylvania wants to use the spatially advanced program to better understand how melanoma cells divide and replicate.
Cohen’s lab is working on making LEVER even more interactive. Undergraduate Drexel student Micheal Koerner is currently utilizing his second co-op to develop a haptic interactive glove that would give the user the ability to touch the data, comprehending the size, shape and texture of the cells with added understanding.
“We can basically create a virtual reality environment where you’re interacting with this data and you’re feeling force feedback as you move your hand with this volume,” Koerner said.
The haptic glove would also serve as a tool with which the user could zoom, pan and scan over the visual data moving in front of them.
“We hope that this new technology will kind of give the biologists a new way of interacting with the data — a more synergistic approach to comprehending it. Allowing them to quantitatively and qualitatively comprehend this data that they have better,” Koerner said.
Cutting-edge software such as LEVER yields the potential to take scientists inside the cells themselves and analyze research in a much more thorough and clerical manner.

